Nearpt2 - Finding Nearest Points in 2D
W. Randolph
Franklin
Rensselaer Polytechnic Institute
Nearpt2 is an online program to find nearest points in 2D. It reads a list of points, and preprocesses them into a data structure. Then it reads a list of query points, and, one by one, finds the closest fixed point to each query point.
Nearpt2 is very fast and very small. One test used vertices of two nonuniform meshes from Mark Shepherd. Processing the 10267 fixed points took about 0.01 CPU seconds on a 1600 MHz Pentium. Querying all 8005 test points took about 0.02 seconds.
Another test used 10,000,000 uniformly randomly generated fixed points and another 10,000,000 query points. The total preprocessing time was 12 CPU seconds on my laptop. The total query time was 65 seconds.
These times exclude reading the points and writing the results, which took about .3 seconds.
Nearpt2 uses linear space and time, and should scale up quite well. I would expect it to handle 100M points.
Algorithm
The data structure is a uniform grid. The preprocessing goes as follows.
- Define N = the number of fixed points.
- Let G = sqrt(N).
- Overlay a GxG grid on the data.
- For each fixed point, insert it into the relevant cell.
Querying with point Q does as follows.
- Find which cell, C, contains Q.
- Find the closest point, if any, in C to Q.
- If C was empty, then sweep out in square rings of diameter 3, 5, 7, ... until a ring is found with at least one point. Find the closest point in that ring to Q.
- Sweep out a little farther to correct for the rings being square. If the ring where the first point was found had radius, sweep out to a ring with radius ceiling(r*sqrt(2)).
Discussion
Nearpt2's expected time is linear in the total number of points. Each query takes constant expected time. An adversary can force a query to take linear time by making the fixed points very uneven. However, I have used this data structure on uneven data from various domains, and never observed this to happen. Note that data that was that uneven would also destroy any the performance of any hierarchical method, like quadtree, because it would require too many levels to be used.
The guiding philosophy is simplicity. That leads to code that is so fast that it can often beat the theoretically better methods like triangulation. Also, the small size means that it scales up better. This idea also scales into higher dimensions, as well as anything does. Finally, this implementation, from creating the directory, thru coding and testing, until this finished web page, took only 6 hours.
The exact value of G is unimportant. Often it has to vary a factor of 3 from the optimum to seriously affect the time.
The program's speed could probably be doubled with some optimization.
Test 1 - Nonuniform Meshes
I tested Nearpt2 on the vertices of two nonuniform meshes of the same region, from Mark Shepherd. Here are the two sets of points. The 10267 red points are fixed and the 8005 green points are queries.
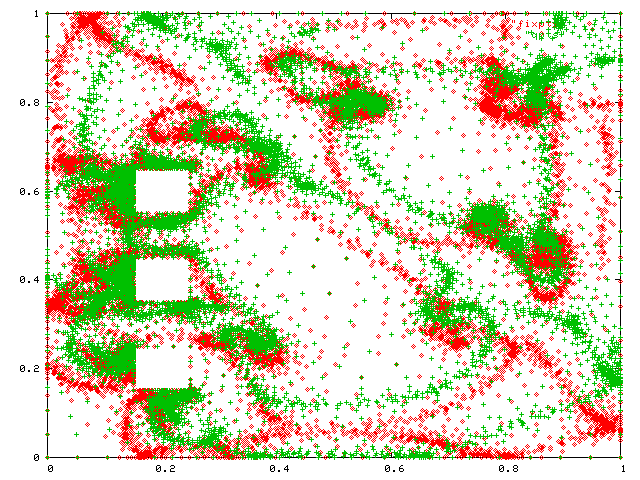
Note how nonuniformly they are distributed. Indeed, the triangles they come from have aspect ratios ranging up to 100:1. The next figure adds an edge from each query to the closest fixed point.
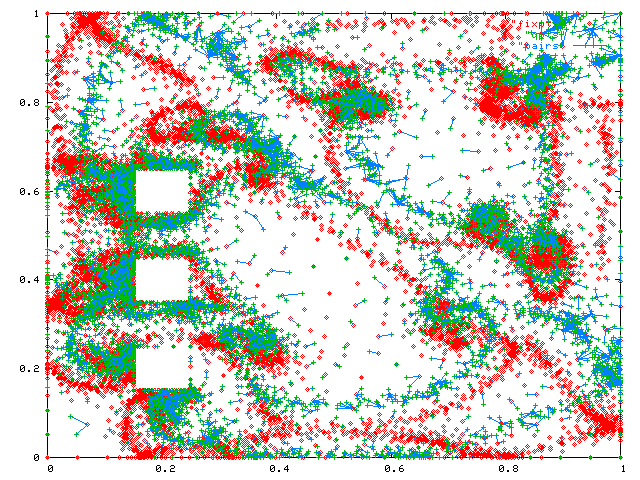
Test 2 - 1,000,000 points each
The fixed and query points were both 1,000,000 uniform independent pseudorandom points. The preprocessing CPU time was 1.1 seconds, and the query time was 5.4 seconds on my laptop. The remaining time was to read 2,000,000 lines and write 3,000,000 lines. The virtual memory was 49MB. The elapsed time including the main program's I/O to find the 1,000,000 nearest points was 41 seconds. (For large runs like this, the number of points was hardcoded into nearpt2.)
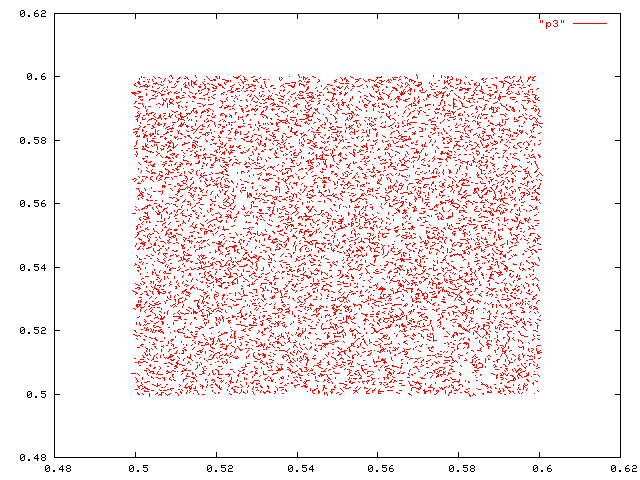
Here's a detail from the plot of closest pairs.
Test 3 - 10,000,000 points each
This test was like before, but with 10,000,000 fixed points and another 10,000,000 query points. The total preprocessing time was 12 CPU seconds on my laptop. The total query time was 65 seconds. The virtual memory was 460MB. The elapsed time including the main program's I/O was 415 seconds.
Implementation
Nearpt2 is written in a C++. Including a main program to do I/O, it's a few hundred lines. My enviromment is SuSE linux 9.0 upgraded with the 2.6.2 kernel, and running g++ or icc, on a 1600MHz IBM Thinkpad T30 laptop. I used icc -O3 to get the times mentioned above.
Nearpt2 reads data from two files fixpts and qpts. Each has one point per line. It requires that the points be in the range [0,1]. It writes to a file called pairs, in the following format, chosen to make plotting in gnuplot easy.
coords of 1st query point coords of closest fixed pt blank line pcoords of 2nd query point coords of closest fixed pt blank line ...
Nearpt2 is freely available for nonprofit research and education. I would appreciate an acknowledgement, and a link to my homepage.
The source is nearpt2.cc. Here are the datasets used for the above images: fixpts and qpts. The resulting output is pairs.